A Step Toward A Quantum Bayesian Network For Rare Disease Diagnosis
Linking Genotype to Phenotype with Metabolomics Data
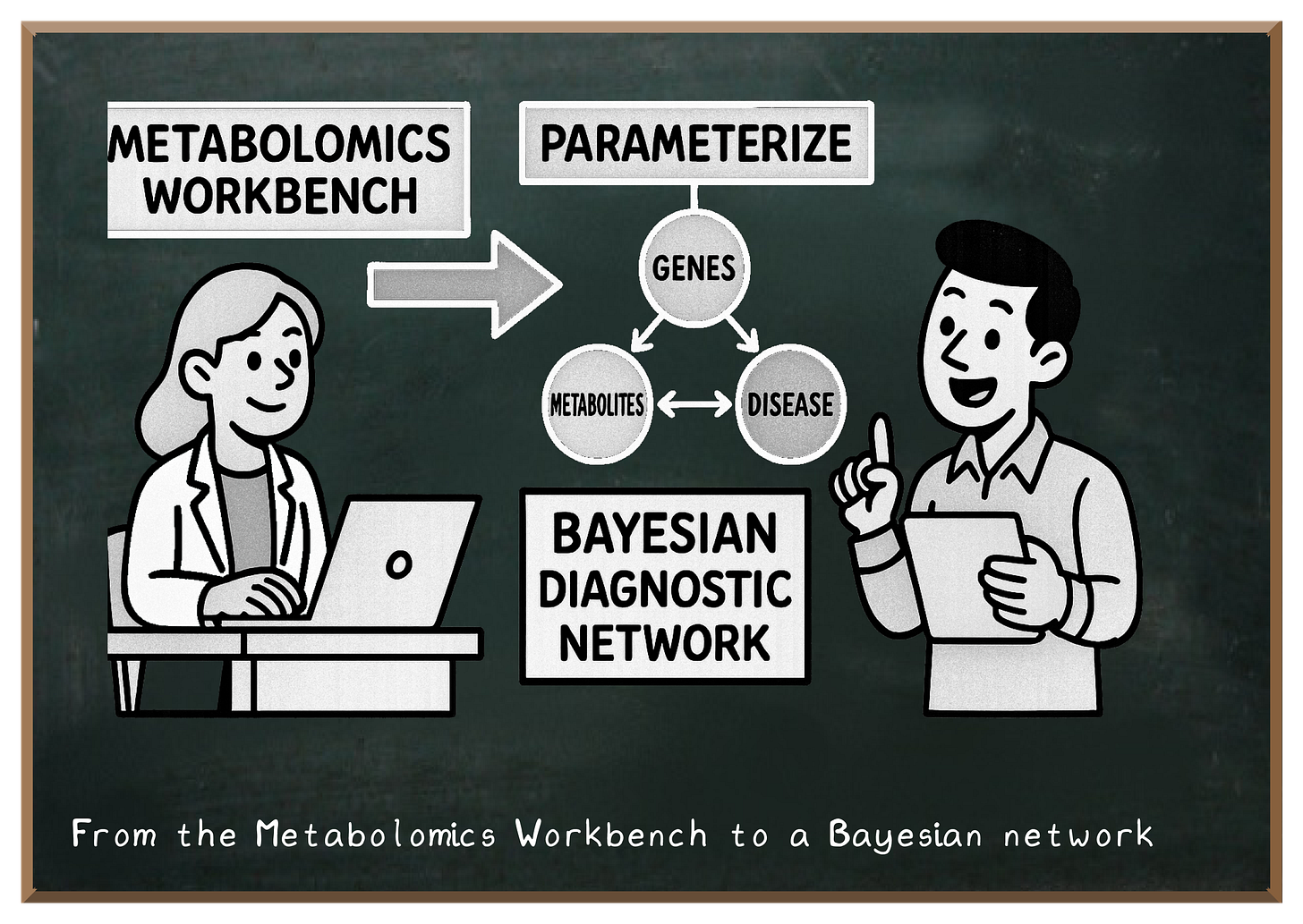
Today's post demonstrates how to leverage **Metabolomics Workbench** datasets to parameterize a **Bayesian diagnostic network**, enabling probabilistic inference between genes, metabolites, and disease states.
From Gene to Metabolite Evidence
The goal is to define the probabilistic relationships between genetic variants, metabolite levels, and clinical phenotypes. For example:
ASS1 deficiency leads to:
Increase of Citrulline
Increase of Aspartate
Decrease of Argininosuccinate
These metabolite changes are measurable in patient lab tests. Using real-world metabolomics datasets allows us to parameterize the conditional probabilities in the diagnostic network.